A recent paper Tensor Product Attention Is All You Need1 grabbed my attention. Over the last year, I have been exploring and investigating ways to reinterpret attention mechanism, mainly for my own edification. What correlations do a transformer really capture? And unsurprisingly, I have been looking at using intuition from the physics of correlated systems.
Firstly, attention mechanism is often written in a mathematically confusing and redundant way in the machine learning literature. The notation is often obfuscated by implementation quirks of matrix multiplications on GPUs. So let’s set up the notation, and simplify.
In the notes below, I will ignore position encoding. RoPE or learnable additive position encodings do not change the foundational mathematical intuitions I am trying to convey here — it is a distraction.
I use for layer index and for head index.
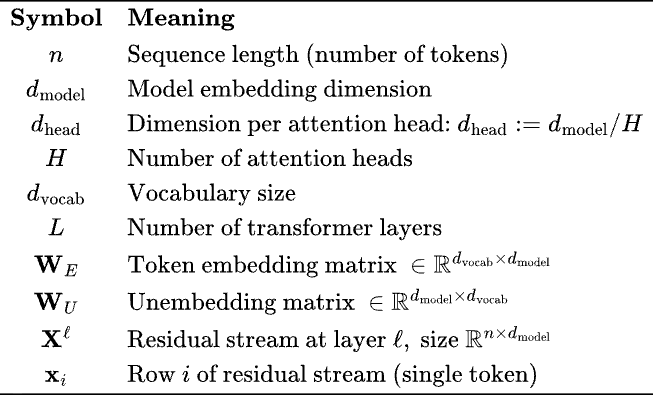
The key quantity is the residual stream, . This matrix is getting transformed by attention and MLP blocks. The embedding dimension is the size of the vector space in which tokens are being embedded.
We need a few other matrices to really explain what’s going on.
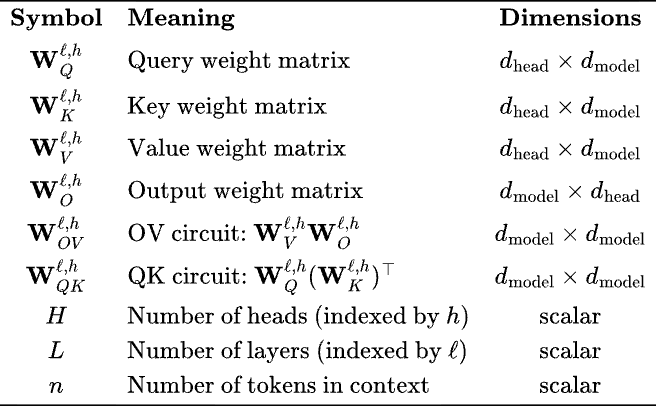
Note that in ML/ AI papers the Query, Value and Key matrices are always written separately, but in essence, we are low-rank decomposing (as product of rectangular matrices) two matrices, . This will be clear when we write attention is terms of these matrices —
The attention operator at layer is a sum over individual attention heads, , with total heads. Note, here I choose to call the operator the net function that returns a vector of same size as — one can choose to add this back to the residual . Some architectures do so, others send it through the MLP operator. There are a lot of different transformer architectures out there in the various LLMs, and for the purpose of this discussion, it’s unimportant. Moreover, the papers have a bewildering range of definitions of what part of is called attention, which is why I bored you with setting up notation. You are welcome.
Note that the number of heads and head dimensions are chosen such that we always have matrices in the above expression.
The only correlation between tokens explored in an transformer is pairwise. The MLP operator acts on the per-token embedding and do not mix and . In the Attention operator term is a normalized weight — and every other token embedding in the context window is getting summed over by this weight multiple by a linear transformation matrix. It is really quite simple.
Well, one may wonder — why only pairwise correlations? And, why only the above functional form for pairwise correlations?
A digression — for physicists like me, any time we see pairwise correlations, we think about Potts model, a generalization of the Ising Model which is perhaps better known. In the q-state Potts model the “spins” are unit vectors that point in q symmetric directions of a hypertetrahedron in q-1 dimensions, see here2. In the classical Potts model these vectors interact only if their “spins” (state) are the same.
Can we draw an analogy with Potts Model? Yes, of course! Well, a paper3 already did a version of it—with a Potts Model where the interactions are not restricted to same “spins” but mix “spins”. It’s an enticing direction to study the dynamics of transformers using such mappings.
OK, end of digression.
The Memory Bottleneck in Modern Transformers
Large language models face a critical scalability challenge: the Key-Value (KV) cache. During autoregressive generation, standard Multi-Head Attention (MHA) stores keys and values for all previously generated tokens, consuming memory that grows linearly with sequence length:
See table to to recall notation. For a model with and processing a token context, this amounts to over 800MB just for the KV cache of a single layer!
The fundamental question is whether we must store the full representation for each token, or whether a more compact factorized representation can capture the essential structure with minimal information loss.
Tensor Decompositions: A Primer
Before diving into Tensor Product Attention (TPA), we need to understand the landscape of tensor decomposition methods. A tensor is simply a multi-dimensional array—scalars are 0-order tensors, vectors are 1st-order, matrices are 2nd-order, and so on.
CP Decomposition (CANDECOMP/PARAFAC)
The most common Tensor Decomposition is probably the CP decomposition.
Definition (CP Decomposition): A third-order tensor has a rank- CP decomposition if it can be written as:
where , , and denotes the outer product.
Element wise, Equivalently, for indices :
The CP decomposition represents a tensor as a sum of rank-1 tensors (outer products of vectors). This is the natural generalization of matrix SVD to higher orders, though unlike SVD, computing the optimal CP decomposition is NP-hard. Yeah, sucks, right?
Tucker Decomposition
Another popular tensor decomposition method is the Tucker Decomposition.
Definition (Tucker Decomposition): A Tucker decomposition factorizes a tensor into a core tensor and factor matrices along each mode: where , , and denotes the mode- product.
More directly, the decomposition is —
The Tucker decomposition generalizes CP by allowing a dense core tensor. Note that the the sizes is obviously within the sizes of the tensor dimensions— a common choice is . When tensor is super-diagonal (non-zero only when all indices are equal), Tucker reduces to CP.
Tensor Train Decomposition
The tensor decomposition most familiar to physicists is probably the tensor train decomposition.
Definition (Tensor Train): A tensor train (TT) or Matrix Product State (MPS) represents a -dimensional tensor as a product of matrices —
where with . The parameters are called bond dimensions or TT-ranks.
This is the same structure used to represent quantum many-body states in physics.
Tensor Product Attention: The Core Claim
Now we arrive at the key contribution of the TPA paper. Instead of storing full query, key, and value matrices, TPA represents them using contextual low-rank factorizations.
Standard Multi-head Attention
For token with embedding , layer and head
We can stack all the heads into matrices, note that now the matrices are not just weights, but weights multiplied by embeddings—
TPA
TPA factorizes the stacked query/key/value matrices as rank- sums of outer products.

Note that the dimensions work out, for clarity —
So for standard MHA, each head independently projects the input—
whereas for TPA, all heads share feature vectors, weighted differently per head,
The Key Idea: Instead of H independent -dimensional vectors (one per head), TPA uses—
- shared feature vectors
- weight vectors — one scalar per head, determining how much each head uses each feature
where , therefore leading to parameter efficiency. Obviously, we have similar things going on for and .
Parameter counts
For MHA, we total number of parameters for queries only (similar for Keys and Values) are
For TPA we have—
- Head factors: matrices of size
- Feature factors: matrices of size
- Total parameters—
Example with typical paper values: , , , :
- MHA: parameters
- TPA: parameters
- TPA uses ~23% of MHA’s parameters
Note: Unlike LoRA which factorizes weights, TPA factorizes activations. This means the factorization is contextual—it depends on the input token . It’s a very interesting idea in how to capture input-dependent structure while maintaining compression!
Memory Reduction
The major advantage claimed by the paper is the memory saving in KV cache. My interest in this paper is beyond this, to study other forms of attention, but it’s useful to note the memory arguments.
From standard MHA we have—
- Store and
- Total:
TPA stores only the factors—
- Store and for keys
- Store and for values
- Total:
The compression ratio is
Concrete example: :
- TPA cache values per token
- MHA cache values per token
so TPA leads to memory reduction! For context window of 100,000 tokens, MHA needs 1.6 GB of memory wheres TPA needs 64 MB of memory! (both per layer)
Connection to MPS
Another way to look at TPA is recasting it as a MPS. Per head, instead of the term in MHA, for TPA we have
We now we are getting somewhere, right? That’s a very different take on the attention matrix capturing token-token correlations!
- Rank indices play the role of bond indices in MPS
- is the bond cotraction
- Low ranks is equivalent to low bond dimension and increased efficiency and high bond dimension leads to more expressiveness
Copy Tensor
We can look at the above expression in terms of copy tensors in Tensor Networks. A copy tensor4 allows for reusing information. For a vector , the copy operation is represented by a diagonal tensor, , the Kronecker delta. In other words, a copy tensor allows a single input to be reused in multiple tensor contractions.
Note what’s happening in TPA! The same input vector is used times for Query, and so on for Key and Value —
Instead of computing H independent projections (standard MHA), TPA computes projections and cleverly recombines them. When , this architecture is much more efficient while maintaining expressiveness of a Tensor Network (outer product).
Few other things…
- The paper shows that TPA is compatible with RoPE embedding. RoPE only acts on the vectors. The keys are pre-rotated and stored, so no rotation is needed during decoding. Only the current query needs to be rotated. Neat!
- Remarkably, standard attention mechanisms are non-contextual variants of TPA! They show that both GQA (Grouped Query Attention) and MQA (Multi-Query Attention) are simply poor man’s version of TPA with being independent of !
I loved the paper. The key lessons:
- Structure matters: Exploiting low-rank structure in attention patterns enables massive compression
- Contextual factorization: Factorizing activations (not weights) is a very interesting concept
- Model performance and memory needs: As with several other work recently, the belief that larger context window either means larger models, or we need to compromise on expressivity of the correlations captured in attention, may be incorrect
As we push toward longer contexts and larger models, principled compression techniques like TPA is a fruitful area of research. The tensor network perspective suggests we’ve only begun to explore the space of possible architectures!
References
- Zhang, Yifan, et al. “Tensor product attention is all you need.” arXiv preprint arXiv:2501.06425 (2025). ↩︎
- Wu, Fa-Yueh. “The Potts Model.” Reviews of modern physics 54.1 (1982): 235. ↩︎
- Rende, Riccardo, et al. “Mapping of attention mechanisms to a generalized Potts Model.” Physical Review Research 6.2 (2024): 023057. ↩︎
- Glasser, Ivan, Nicola Pancotti, and J. Ignacio Cirac. “From probabilistic graphical models to generalized tensor networks for supervised learning.” IEEE Access 8 (2020): 68169-68182. ↩︎